As part of its Pathology at Life Speed (PALS) webinar series, Proscia hosted an event earlier this month focused on the promise of foundation models in driving pathology innovation. This session provided a deep dive into one of the largest foundation models for pathology AI development, with practical strategies for data scientists and AI developers to leverage these models for transformative impact on drug discovery and development. If you missed the live session, you can catch the full recording here. Featured speakers from the webinar include:
- Julianna Ianni, Ph.D. — Vice President, AI Research & Development, Proscia
- Corey Chivers, Ph.D. — Senior AI Scientist, Proscia
- Zelda Mariet, Ph.D. — Co-founder and Principal Research Scientist, Bioptimus
Here are a few highlights from the webinar:
Computational pathology’s strategic role in precision medicine
Dr. Julianna Ianni of Proscia kicked off the discussion by underscoring the rising importance of computational pathology in life sciences organizations’ AI strategies. Computational pathology is revolutionizing therapeutic R&D by enabling precise analysis of hundreds of thousands of cells per slide, streamlining decision-making, driving innovation, and ultimately bringing new medicines to patients faster.
Julianna shared how organizations are developing their own novel pathology AI models to accelerate AI development cycles, retain proprietary data, and strengthen their competitive advantage. For example, during their recent AI Day, BioNTech highlighted building and deploying histology AI to enhance the speed and accuracy of tissue labeling as one of their five core strategies for creating an AI-first immunotherapy platform.
Beyond driving efficiencies, the rise of precision medicine approaches and the rapid growth of next-generation therapeutics have elevated computational pathology to a strategic enabler. Julianna emphasized that timely access to advanced diagnostic tools is crucial for these treatments to reach and benefit patients effectively. Computational pathology is driving this paradigm shift by enabling the development of algorithms that detect oncology biomarkers in digitized tissue biopsies. This provides rapid, actionable disease insights that empower clinicians to make more informed decisions and ultimately improve patient outcomes.
The growth of computational pathology extends beyond the pharmaceutical industry. Julianna highlighted how this expansion is also evident in scientific literature, showcasing advancements in academia, clinical applications, and therapeutic R&D. Notably, over 70% of publications in this field have been released in the past five years, underscoring a rapidly accelerating trend.
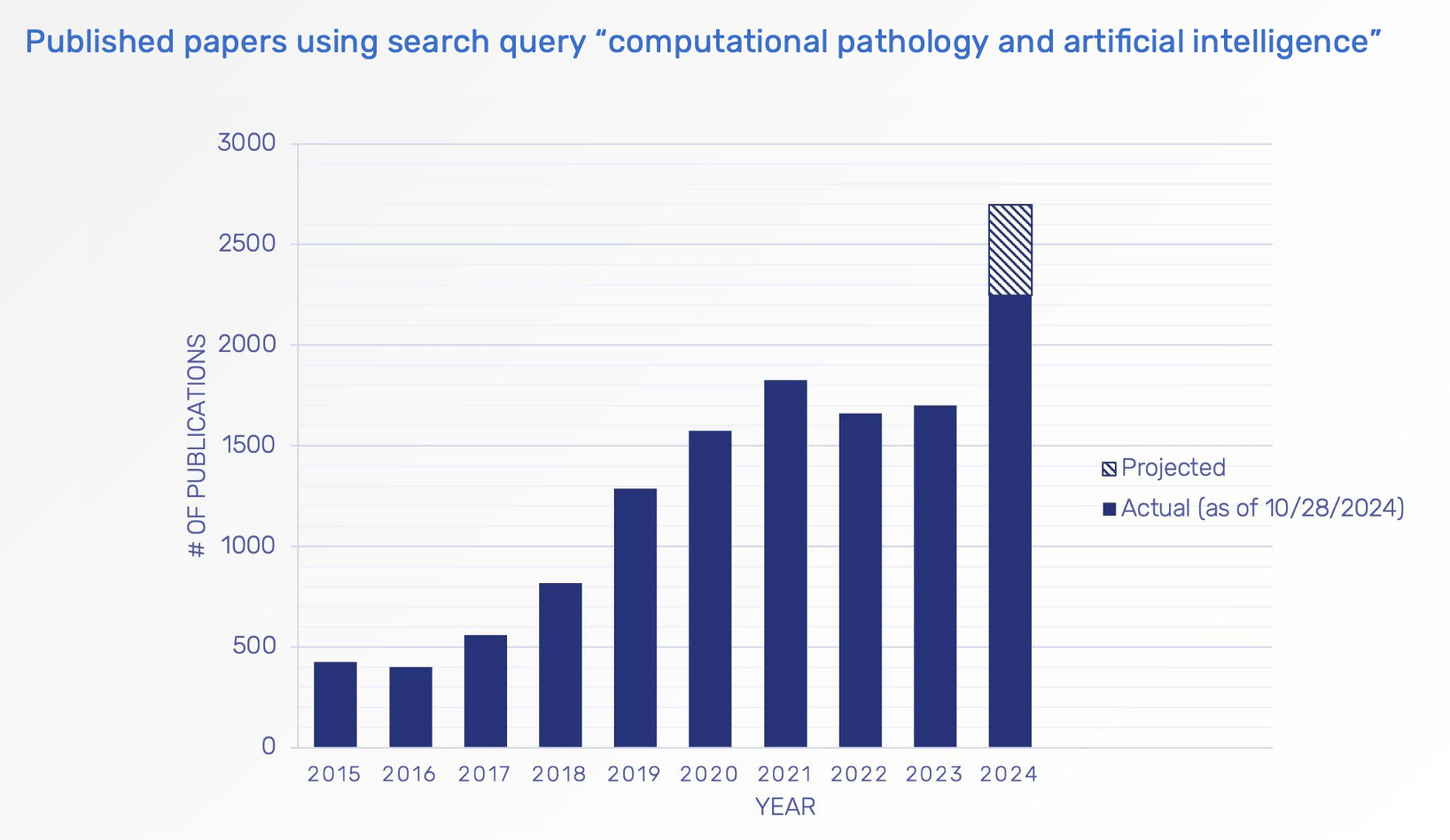
Chart presented by Julianna Ianni, Ph.D., during webinar
Julianna also highlighted how digital pathology platforms have advanced significantly over the past two decades, creating a solid foundation for computational pathology to make a tangible clinical impact. These platforms provide life sciences organizations with the tools necessary to analyze images at scale, a critical capability in today’s data-driven landscape. As she noted, “There is no way to build or use AI if all of your slides are sitting in file cabinets or on hard drives on people’s desks.”
In addition, foundation models have emerged as a transformative technology, enabling the faster and more efficient development of pathology AI models—setting the stage for the webinar’s next topic.
How foundation models are reshaping pathology AI development
Dr. Zelda Mariet of Bioptimus provided a background on foundation models, explaining that they typically involve billions of parameters and must be trained on massive datasets. She asked the audience to think of them as the histopathology equivalent of GPT-style models: they are capable of understanding the intricate structures within whole slide images (WSIs) without requiring large amounts of labeled data. These models serve as a “foundation” for downstream applications, enabling researchers to leverage their capabilities with minimal additional training.
In histopathology, Zelda explained, the data scale is unparalleled. A single WSI contains a staggering 100,000 x 100,000 pixels—an immense wealth of biological information. However, the challenge lies in the scarcity of labeled data; most WSIs lack the detailed annotations required by traditional machine learning approaches. This is where foundation models shine: they learn from the structure and patterns within the data itself, eliminating the reliance on annotations.

Slide presented by Zelda Mariet, Ph.D., during webinar
Zelda explained the two primary methods that power the learning process of foundation models in histopathology:
- Contrastive Learning
The model learns by distinguishing between similar and dissimilar samples. For instance, it examines patches of a single tissue slide and determines whether they originate from the same tissue region. This method allows the model to understand biological structures and relationships within the slide. - Generative Learning (Masked Modeling)
Here, the model “fills in the blanks.” Portions of an image are masked, and the model is tasked with reconstructing the missing sections. This approach encodes fundamental biological structures, enabling the model to recognize plausible versus implausible reconstructions.
Through these techniques, foundation models create embeddings—compact representations of complex WSIs—that retain essential biological information in a much smaller data space.
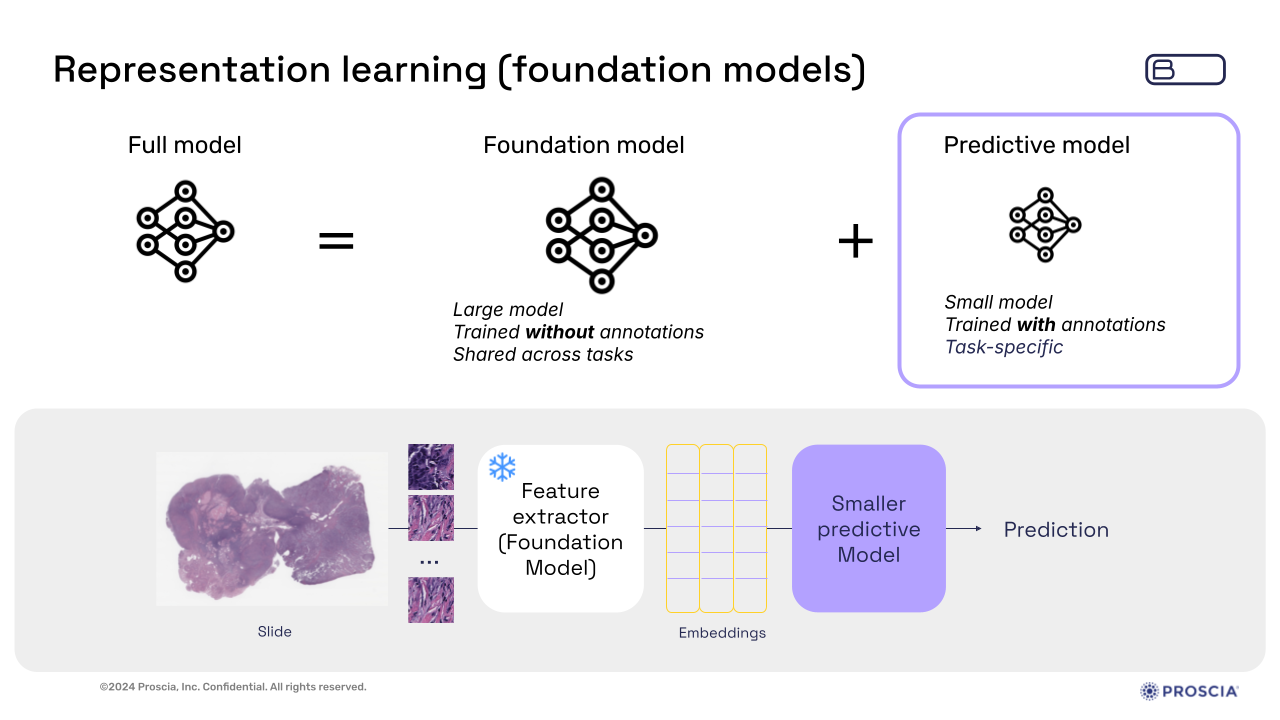
Slide presented by Zelda Mariet, Ph.D., during webinar
Bioptimus’s flagship foundation model, H-optimus-0, exemplifies the power of this technology. Zelda explained how the model is trained on over 600,000 slides and data from 200,000 cancer patients, representing a monumental step forward in histopathology AI. With over one billion parameters and advanced architecture, H-optimus-0 has demonstrated superior accuracy in tasks like cancer subtyping and biomarker detection.
Available on Hugging Face, H-optimus-0 is open source. It’s also now integrated into Proscia’s Concentriq platform through Concentriq Embeddings, right where an organization’s pathology data is stored, enriched, and analyzed.
Zelda also provided one state-of-the-art application for H-optimus-0 involving gene expression prediction from histology slides. Despite limited data availability for gene expression, H-optimus-0 has shown remarkable accuracy in predicting this information solely from WSIs, and is currently leading the HEST-Benchmark run by the Mahmood Lab out of Harvard Medical School. This capability holds immense potential for advancing research in precision medicine and drug discovery.
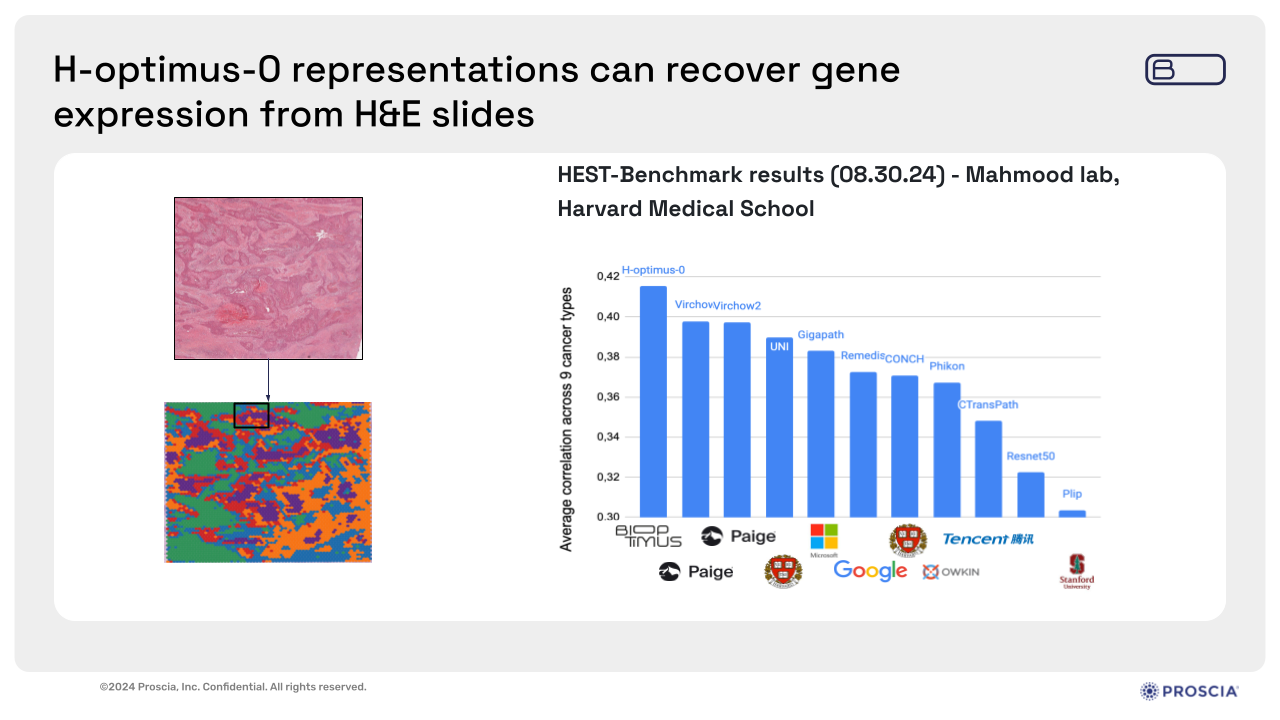
Slide presented by Zelda Mariet, Ph.D., during webinar
Practical strategies for leveraging foundation models for pathology AI development
It’s clear that foundation models hold transformative potential for digital histopathology, but their implementation is not without challenges. From inefficient workflows to high computational costs, and the complexity of integrating multiple models, Dr. Corey Chivers of Proscia reviewed these challenges and how they often slow progress and limit innovation.
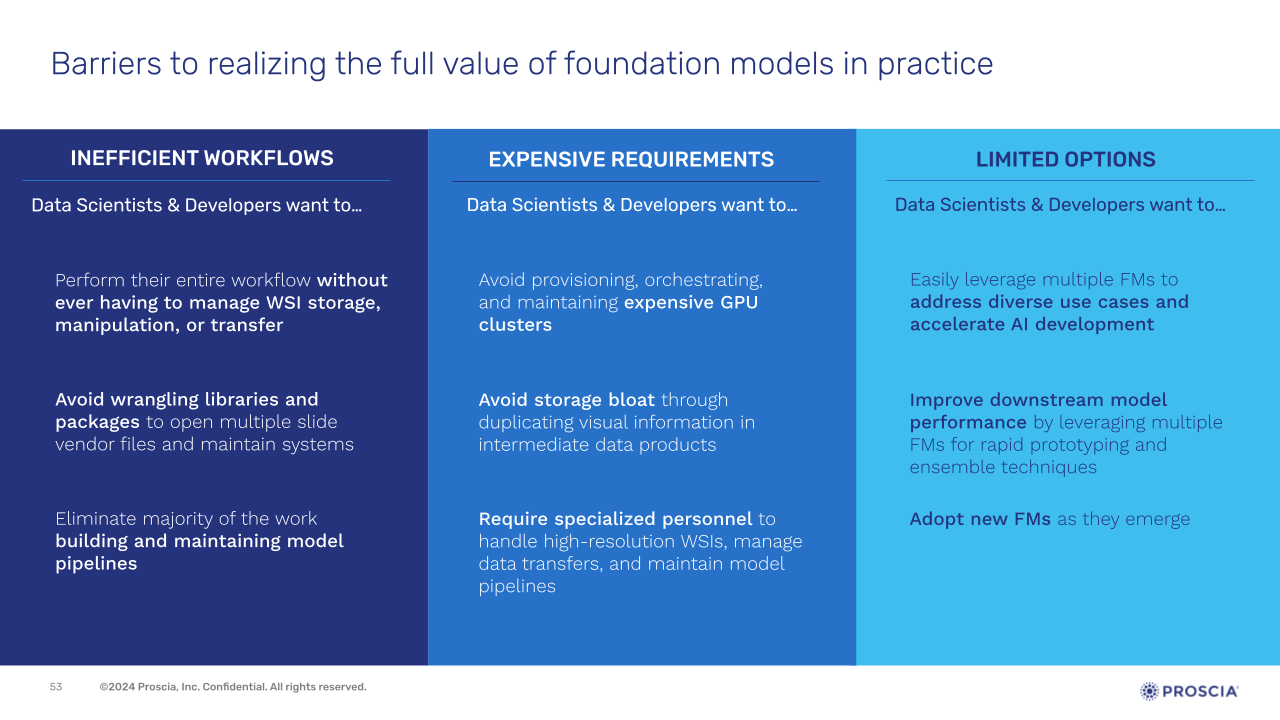
Slide presented by Corey Chivers, Ph.D., during webinar
Corey provided an overview of a few recent independent foundation model benchmarking research to demonstrate the importance of a multi-model strategy—a concept central to Proscia’s Concentriq Embeddings. These studies have demonstrated that no single foundation model consistently outperforms others across all tasks and datasets, and that ensembles of complementary models can outperform the best-performing foundation model in approximately two-thirds of tasks.
These results highlight the substantial benefits of having multiple foundation models available to leverage, whether to rapidly prototype and find the right model for your downstream task faster, or to combine the strengths of multiple models to achieve more accurate and robust predictions.
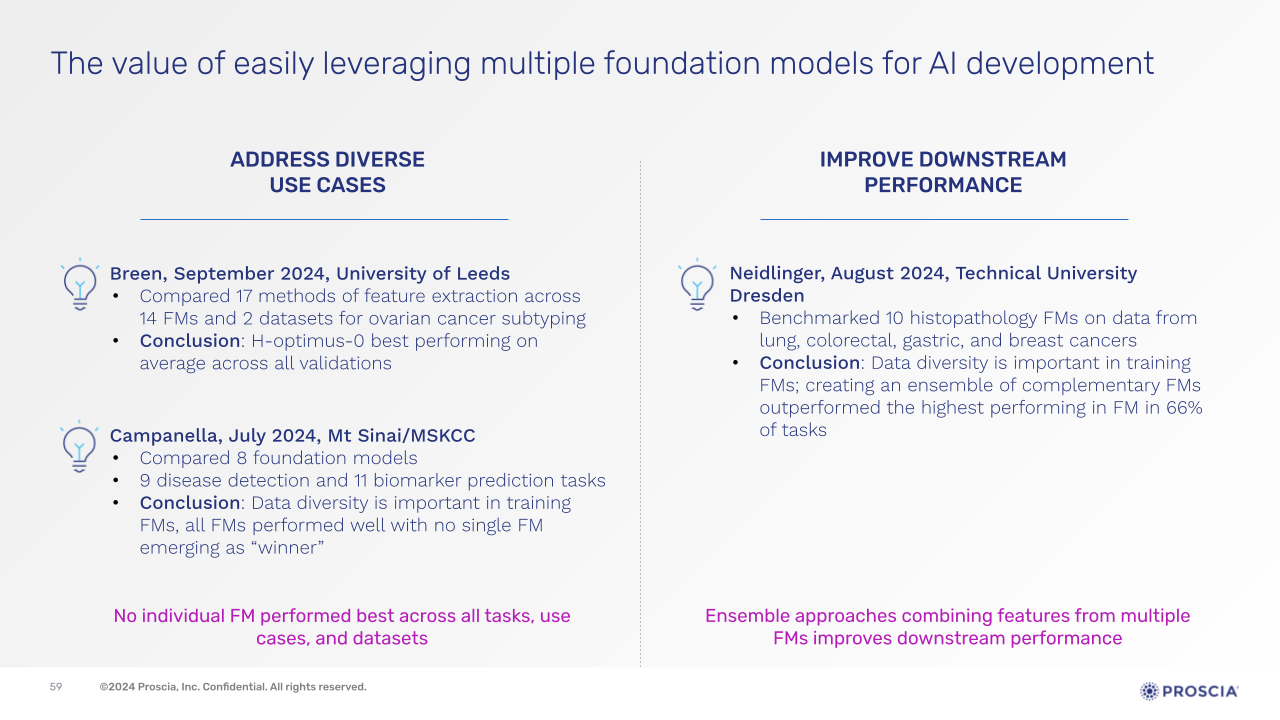
Slide presented by Corey Chivers, Ph.D., during webinar
In response to the challenges and opportunities presented with leveraging foundation models, Proscia has developed Concentriq Embeddings, a solution designed to remove these barriers and streamline AI model development for life sciences organizations.
Corey explained how Concentriq Embeddings empowers data scientists and AI developers to harness foundation models more efficiently and effectively, so they can:
- Accelerate Therapeutic R&D: Develop AI models faster to drive impactful R&D processes and insights. For instance, in an internal case study using Concentriq Embeddings on a consumer-grade laptop, Proscia successfully trained and evaluated 80 breast cancer biomarker prediction models in less than 24 hours.
- Optimize AI Model Performance: Use multiple foundation models to quickly find the best fit for datasets and applications, as well as leverage ensemble approaches.
- Reinvest Computational Savings: Eliminate low-value tasks required to set up and manage foundation models and instead allocate resources towards meaningful AI transformation.
Corey provided a demonstration of how Concentriq Embeddings works in practice, which is available through registering for the webinar here.
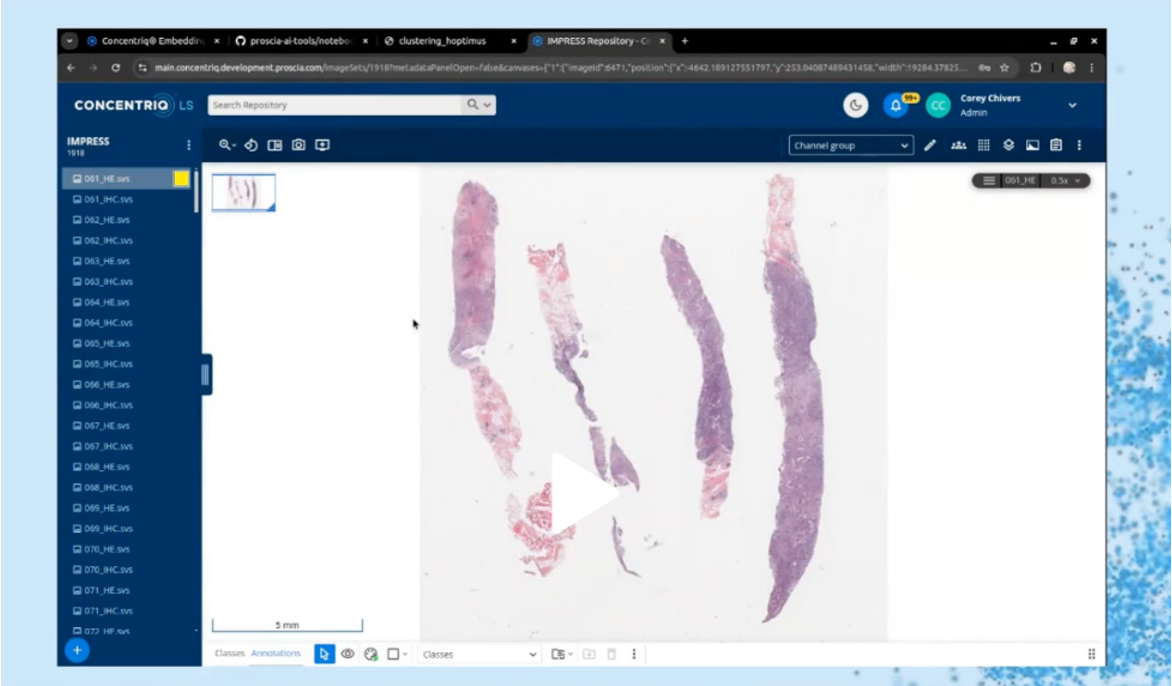
Corey Chivers, Ph.D. provided a demonstration of Concentriq Embeddings
Conclusion: The path forward with foundation models
Foundation models represent a paradigm shift in how we approach computational pathology and AI-driven drug discovery and development. This webinar demonstrated their transformative potential, not only in accelerating R&D but also in reshaping precision medicine. With tools like Concentriq Embeddings, life sciences organizations can overcome barriers to implementation, unlocking new possibilities in therapeutic innovation and patient care.
As the technology continues to evolve, foundation models are poised to become the cornerstone of pathology AI development, driving faster discoveries, better diagnostics, and ultimately, improved patient outcomes.
Missed the live session? Watch the full webinar recording here to explore these topics further.
