The editors of Nature Methods unanimously recognized spatial transcriptomics as Method of the Year 2020. Rightfully so, the exciting field of spatial biology methods can illuminate single-cell heterogeneity and define cell types while also retaining spatial information. According to Nature Methods, “This maintenance of spatial context is crucial for understanding key aspects of cell biology, developmental biology, neurobiology, tumor biology and more, as specialized cell types and their specific organization are crucially tied to biological activity and remain poorly explored on the scale of whole tissues and organisms.” Today, organizations across the world are taking advantage of spatial biology techniques to one day generate complete maps of large and complex tissues such as the human brain. To do so, many scientists start with a future-ready, robust digital pathology platform to view, analyze, and collaborate on Whole Slide Images (WSI) as well as take advantage of the advances in computational biology to get more out of their data.
Why Multiplexed Tissue Imaging within Spatial Biology?
Imaging has been the tool of choice for analyzing spatial organization and better understanding disease progression and treatment options at the tissue level. More specifically, fluorescent imaging is one of the most widely used techniques in life sciences research, from cancer research and immunology to neuroscience and more. Both fluorescent imaging – and more specifically, highly multiplexed imaging, an emerging technique in spatial biology – has provided unprecedented insights into disease states. Most recently, Nature Methods highlighted the value of multiplexed tissue imaging for enabling researchers to gather rich information to complement data gathered using traditional approaches. With immunofluorescence enabling high spatial resolution over large regions, this method is desirable for imaging the location and highlighting an abundance of specific proteins of interest.
A Case Study of the Power of Harnessing Spatial Biology & Digital Pathology in Cancer Immunotherapy
In his article in The Pathologist, Ori Zelichov talks about the potential of spatial biology and artificial intelligence (AI) for analyzing the tumor microenvironment (TME) of novel markers. With the help of advanced imaging algorithms, scientists and clinicians have the potential to analyze Whole Slide Images (WSI) for a comprehensive description of the tumor and the TME. Such an analysis could also help identify the subtle immune signatures that distinguish between immunotherapy responders and non-responders. This would help to improve treatment selection of patients in both clinical trials and in the real world.
Zelichov talks about another use case as well: patients, who upon analysis of TME, are not good candidates for immunotherapy and thus lack effective treatment options. In this case, there is a potential to use the power of spatial biology and artificial intelligence (AI) to combine immunotherapy with TME targeting agents (such as those that can potentially reprogram immunosuppressive cells to overcome resistance mechanisms in the TME and to enhance antitumor immunity).
In Zelichov’s opinion, new drugs targeting the TME could potentially help immuno-oncology to better treat unaddressed patient populations with unresponsive tumors. In other words, understanding “hot” and “cold” spatial arrangements can translate into actionable treatment decisions. This is potentially very good news. Increasingly, the use of advanced AI tools for in-depth tissue analysis could become a leading way to learn more about the TME, understand drug mechanisms of action, and guide the discovery and development of a new class of immuno-oncology drugs for non-responding immunotherapy patients.
How Proscia’s Concentriq for Research empowers scientists to accelerate their fluorescence image-based research
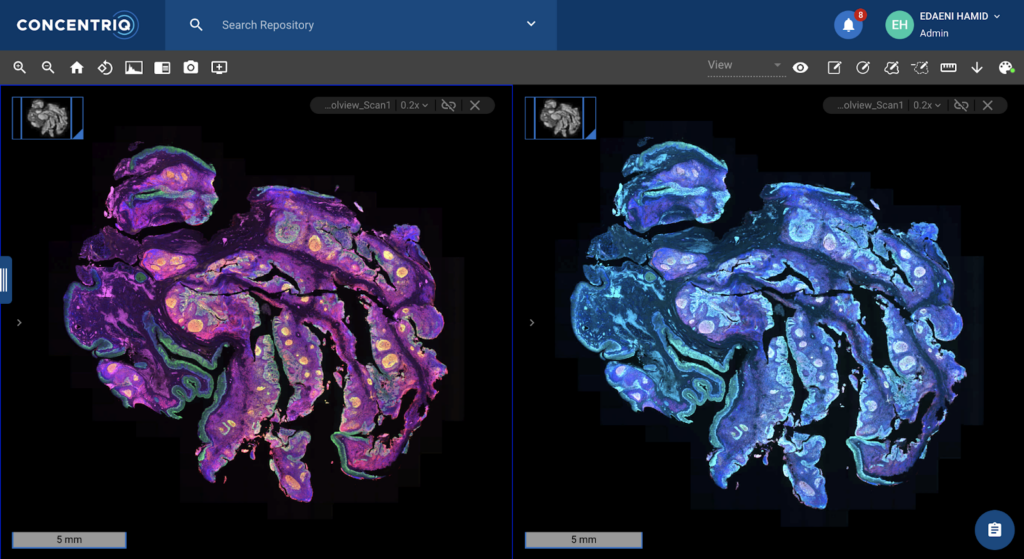
To support spatial biology research in the life sciences and provide tools to assist with breakthrough innovations, Concentriq for Research supports fluorescence imaging in the same mature enterprise-grade platform that has enabled image-based research for some of the largest global teams. Key fluorescence image capabilities available in Concentriq for Research include:
- Scanner-agnostic platform: Concentriq for Research accepts and supports a wide variety of fluorescent image formats from various vendors. Users can share repositories and groups with collaborators who are then able to upload images into these repositories
- An intuitive workflow for fluorescent image viewing: Users are able to compare multiple fluorescent and brightfield images side-by-side in Concentriq for Research’s multiview. Additionally, biomarker names – separable from channel names – are visible alongside the images, giving researchers additional context about the images they’re analyzing. Other, general interface enhancements – including an easy-to-use intensity adjuster – deliver a simplified and efficient workflow for researchers who review fluorescent images regularly.
- Easy access to fluorescent images and fields alongside brightfield and other imaging data: Users are able to leverage the power of Concentriq for Research’s robust global search functionality on fluorescent images. The images are searchable by biomarkers, allowing the organization to easily access their past and present data. This metadata is organized and managed alongside other pathology data, making it easier than ever to bring the power of fluorescent imaging to a broader body of R&D.
- Standardized fluorescent images and data management across the organization: Support for fluorescence in project templates and metadata field libraries encourages the collection of consistent data across studies. Concentriq for Research allows admin users to create and manage a library of metadata fields for the organization, specifying what the metadata field applies to (such as folder, slide, or annotation) and in what form the data is captured (such as dropdown, checkbox, date, number, and text). The platform also enables users to create metadata templates, which can be applied to studies of similar types repeatedly. By expanding this platform functionality to fluorescent images, scientists can better incorporate this imaging modality alongside other pathology data.
Check out the product tour hosted by Nathan Buchbinder and Edaeni Hamid to learn more about the latest features of Concentriq for Research 3.7.
Listen to Andrew Fisher, Principal Scientist in the Translational, Bioinformatics, Digital Pathology Group at Bristol-Myers Squibb to discuss how they are using Concentriq platform to advance the field of pathology and our collective understanding of tissue via the spatial transcriptomics, computational pathology, and related disciplines involving tissue imaging.
